Web-scraping is essentially the task of finding out what input a website expects and understanding the format of its response. For example, Recovery.gov takes a user’s zip code as input before returning a page showing federal stimulus contracts and grants in the area.
This tutorial will teach you how to identify the inputs for a website and how to design a program that automatically sends requests and downloads the resulting web pages.
Pfizer disclosed its doctor payments in March as part of a $2.3 billion settlement – the largest health care fraud settlement in U.S. history – of allegations that it illegally promoted its drugs for unapproved uses.
Of the disclosing companies so far, Pfizer’s disclosures are the most detailed and its site is well-designed for users looking up individual doctors. However, its doctor list is not downloadable, or easily aggregated.
So we will write a scraper to download Pfizer’s list and record the data in spreadsheet form. The example code is written in Ruby and uses the Nokogiri parsing library.
You may also find Firefox’s Firebug plugin useful for inspecting the source HTML. Safari and Chrome have similar tools built in.
Scouting the Parameters
Pfizer’s website presents us with a list of health care providers – but not all of them at once. There are links to lists that show payees by the first letter of their names, with links to jump deeper into the lists, as each page only shows 10 entries at a time.
Other than the actual list entries themselves, Pfizer’s website looks pretty much the same no matter where in the list you are. It’s safe to assume that there’s one template with a slot open for the varied list results.
How does the site know what part of the list to serve up? The answer is in your browser URL bar. Clicking on the link for ‘D’ gives us this address:
http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?enPdNm=D
For the letter ‘E’, the address is this:
http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?enPdNm=E
So the only difference in the address is the letters themselves. Instead of physically clicking the links for each letter, you can just change them in the address bar.
If you click through other pages of the list, you’ll see that the one constant is this:
http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?enPdNm=E
With this as the base address, what follows the question mark are the parameters that tell Pfizer’s website what results to show. For the list of alphabetized names, the name of the is enPdNm.
So just as it’s possible to skip the link-clicking part to navigate the list, it’s possible to automate the changing of these parameters instead of manually typing them.
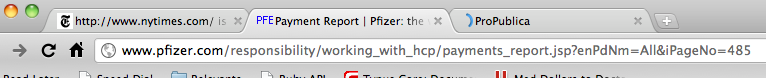
How long is this list anyway?
The Pfizer website gives us the option of paging through their entire list, from start to finish, 10 entries at a time. Clicking on the “Last” link reveals a couple of things about the website:
- There are 4,850 entries total.
- There are 485 pages total. You can see that even without dividing 10 into 4,850 by looking at the page’s url. There’s a parameter there called iPageNo and it’s set to 485. Changing this number allows you to jump to any of the numbered pages of the list.
Data Structure
Upon closer inspection of the page, you’ll notice that the names in the left-most column don’t refer to health care providers, but the person to whom the check was made out. In most cases, it’s the same name of the doctor; in others, it’s the name of the clinic, company, or university represented by the doctor.
The right column lists the actual doctor and provides a link to a page that includes the breakdown of payments for that doctor.
In this list view, however, the payment details are not always the same as what’s shown on each doctor’s page. Click on an doctor’s name to visit his/her page. In your browser’s address bar, you’ll notice that the website incorporates a new parameter, called “hcpdisplayName“. Changing its value gets you to another doctor’s page. Caveat: after scraping the site, you’ll see that this page does not always include just one doctor. See the caveats at the end of this section.
The Scraping Strategy
I’ll describe a reasonably thorough if not quick scraping method. Without knowing beforehand the details of how Pfizer has structured their data, we can at least assume from scouting out their site’s navigation that:
- Every entity is listed once in the 485 pages.
- Every doctor who’s worked with Pfizer in a disclosed capacity is connected to at least one of these entities.
So, our scraping script will take the following steps:
- Using the parameter enPdNm, Iterate through each pages 1 to 485 and collect the doctors’ names and page links
- Using the links collected in step 1, visit each doctor’s page – using the parameter hcpdisplayName – and copy the payment details.
Downloading the Lists
We can write a script to help us save the list pages to our hard drive, which we’ll read later with another script.
# Call Ruby's OpenURI module which gives us a method to 'open' a specified webpage require 'open-uri' # This is the basic address of Pfizer's all-inclusive list. Adding on the iPageNo parameter will get us from page to page. BASE_LIST_URL = 'http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?enPdNm=All&iPageNo=' # We found this by looking at Pfizer's listing LAST_PAGE_NUMBER = 485 # create a subdirectory called 'pfizer-list-pages' LIST_PAGES_SUBDIR = 'pfizer-list-pages' Dir.mkdir(LIST_PAGES_SUBDIR) unless File.exists?(LIST_PAGES_SUBDIR) # So, from 1 to 485, we'll open the same address on Pfizer's site, but change the last number for page_number in 1..LAST_PAGE_NUMBER page = open("#{BASE_LIST_URL}#{page_number}") # create a new file into to which we copy the webpage contents # and then write the contents of the downloaded page (with the readlines method) to this # new file on our hard drive file = File.open("#{LIST_PAGES_SUBDIR}/pfizer-list-page-#{page_number}.html", 'w') # write to this new html file file.write(page.readlines) # close the file file.close # the previous three commands could be condensed to: # File.open("#{LIST_PAGES_SUBDIR}/pfizer-list-page-#{page_number}.html", 'w'){|f| f.write(page.readlines)} puts "Copied page #{page_number}" # wait 4 seconds before getting the next page, to not overburden the website. sleep 4 end
And that’s it for collecting the webpages.
Parsing HTML with Ruby’s Nokogiri
A webpage is nothing more than text, and then more text describing how its all structured. To make your job much easier, you’ll want to use a programming library that can quickly sort through HTML’s structure.
Nokogiri is the recommended library for Ruby users. Here’s an example of it in use and how to install it. I describe its use for some basic parsing in my tutorial on reading data from Flash sites.
Data-heavy websites like Pfizer’s pull data from a database and then use a template to display that data. For our purposes, that means every page will have the same HTML structure. The particular datapoint you want will always be nested within the same tags.
Finding a webpage’s structure is best done through your browser’s web development tools, or plugins such as Firefox’s immensely useful Firebug. Right clicking on a doctor’s name (and link) in Google Chrome will bring up a pop-up menu.
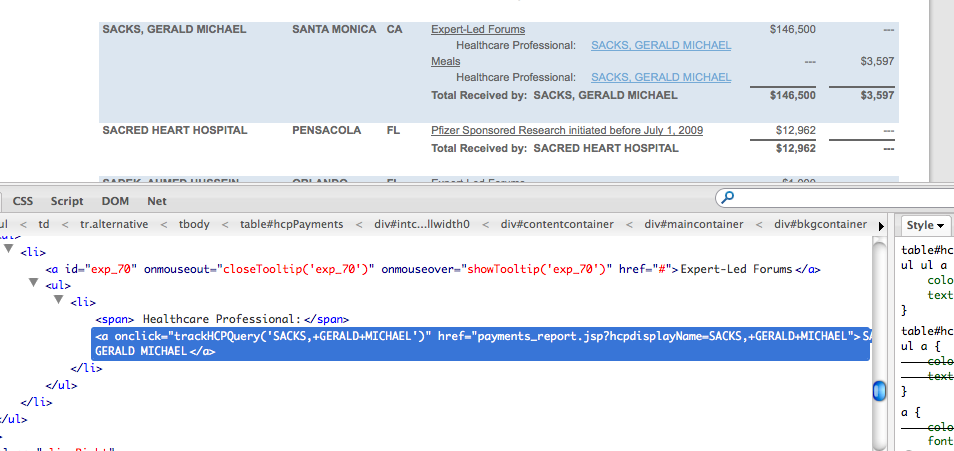
The doctor’s name is wrapped up in an <a> tag, which is what HTML uses to determine a link. The ‘href’ bit is the address of that doctor’s page.
So these <a> tags contain all the information we need right now. The problem is that there are non-doctor links on every page, too, and we don’t want those.
Here’s a quick, inelegant solution: Use Nokogiri to select all of the links and then filter out the ones we don’t want. Looking at our browser’s web inspector, we see that every doctor link has an href attribute that includes the parameter hcpdisplayName:
http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?hcpdisplayName=SACKS,+GERALD+MICHAEL
So using the Nokogiri library (here’s a primer to its basic syntax), this is how you open a page:
require 'rubygems' require 'nokogiri' require 'open-uri' url = 'http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?enPdNm=All&iPageNo=1' page = Nokogiri::HTML(open(url)) # page now contains the HTML that your browser would have read had you visited the url with your browser
The following code example assumes you downloaded the Pfizer list pages as described in the previous step to your hard drive into the specified directory.
The variable n_page contains the same thing that the open command gave us in the pre-step, but in a special Nokogiri construct.
Nokogiri’s library now lets us navigate the HTML in a more orderly fashion than if it were plain text. So, to grab all the links (<a> tags), we use Nokogiri’s css method, which returns an array of all those links:
links = page.css('a')
puts links.length # this prints out '201'
We know there aren’t 201 doctor links on this page. So let’s filter this collection of links using Ruby’s map and select methods.
Remember that each “link” contains more than an address. Each member of the links array is a Nokogiri object that contains numerous properties. The only one we want is ‘href‘. This can be done by calling Ruby’s map method, which will return an array of just what was in each corresponding link’s href attribute (i.e. the array will contain the same number of entries as before: 201).
hrefs = links.map{ |link| link['href'] }
The select method will keep members of a collection that meet a certain true/false test. We want that test to be: does the link’s href attribute contains the word hcpdisplayName?
doc_hrefs = hrefs.select{ |href|
href.match('hcpdisplayName') != nil
}
doc_hrefs = doc_hrefs.uniq
The match method returns nil – i.e. Ruby’s term for ‘nothing/nada/zilch’ – if there was no match. So, we want all href’s that don’t return nil.
We finish by calling the uniq method, which returns only unique values. Remember that a doctor’s name can be repeated on any given list page, depending on the kind of payment records he/she is listed under. We only have to worry about finding the one link to his/her individual page.
You can narrow the number of links to consider by using Nokogiri’s css method to limit the links to just the third column in the table. If you view the page’s source (made easier by using Firebug), then you’ll see that the payments table has an id of “hcpPayments.” Use the CSS pseudo-class nth-child to specify the fourth column.
The full script to read our previously downloaded list pages and write the doctor page URLs to a text file is:
require 'rubygems'
require 'nokogiri'
LIST_PAGES_SUBDIR = 'pfizer-list-pages'
DOC_URL_FILE = "doc-pages-urls.txt"
# Since a doctor's name could appear on different pages, we use a temporary array
# to hold all the links as we iterate through each page and then call its uniq method
# to filter out repeated URLs before writing to file.
all_links = []
# This Dir.entries method will pick up the .html files we created in the last step
Dir.entries(LIST_PAGES_SUBDIR).select{|p| p.match('pfizer-list-page')}.each do |filename|
puts "Reading #{filename}"
n_page = Nokogiri::HTML(open("#{LIST_PAGES_SUBDIR}/#{filename}"))
all_links += n_page.css('table#hcpPayments td:nth-child(4) a').map{|link| link['href'] }.select{|href| href.match('hcpdisplayName') != nil}
end
File.open(DOC_URL_FILE, 'w'){|f| f.puts all_links.uniq}
If you open the doc-pages-urls.txt that you just created, you should have a list of more than 4,500 (relative) urls. The next step will be to request each of these pages and save them to your hard drive, which is what we did earlier, just with a lot more pages.
For beginning coders: There’s a couple of new things here. We call the match method using a regular expression to match the doctor’s name that’s used as the parameter. If you’ve never used regular expressions, we highly recommend learning them, as they’re extremely useful for both programming and non-programming tasks (such as searching documents or cleaning data).
Because this script has a long execution time, we’ve added some very rudimentary error-handling so that the script doesn’t fail if you have a network outage. In the rescue block, the script will sleep for a long period before resuming. Read more about error-handling.
require 'open-uri'
# This is the basic address of Pfizer's all-inclusive list. Adding on the iPageNo parameter will get us from page to page.
BASE_URL = 'http://www.pfizer.com/responsibility/working_with_hcp/'
DOC_URL_FILE = "doc-pages-urls.txt"
DOC_PAGES_SUBDIR = 'pfizer-doc-pages'
Dir.mkdir(DOC_PAGES_SUBDIR) unless File.exists?(DOC_PAGES_SUBDIR)
File.open(DOC_URL_FILE).readlines.each do |url|
# using regular expression to get the doctor name parameter to name our local files
doc_name=url.match(/hcpdisplayName=(.+)/)[1]
begin
puts "Retrieving #{url}"
page = open("#{BASE_URL}#{url}")
rescue Exception=>e
puts "Error trying to retrieve page #{url}"
puts "Sleeping..."
sleep 100
retry # go back to the begin clause
else
File.open("#{DOC_PAGES_SUBDIR}/pfizer-doc-page--#{doc_name}.html", 'w'){|f| f.write(page.readlines)}
ensure
sleep 4
end
end
Reading Every Doctor Page
This step is basically a combination of everything we’ve done so far, with the added step of writing the data for each doctor in a normalized format. So:
- For each address listed in the ‘doc-pages-urls.txt’ file, retrieve the page and save it.
- Read each of the saved pages and parse out the tables for the relevant data.
- Delimit, normalize and print the data to a textfile
Normalizing the Data
We’ll be storing Pfizer’s data in a single text file, and so we want every kind of payment to have its own line. This means we’ll be repeating some fields, such as the doctor’s name, for each payment. It may look ugly and redundant, but it’s the only way to make this data sortable in a spreadsheet. Here’s a diagram of how the web site information will translate into the text-file. Click to enlarge.
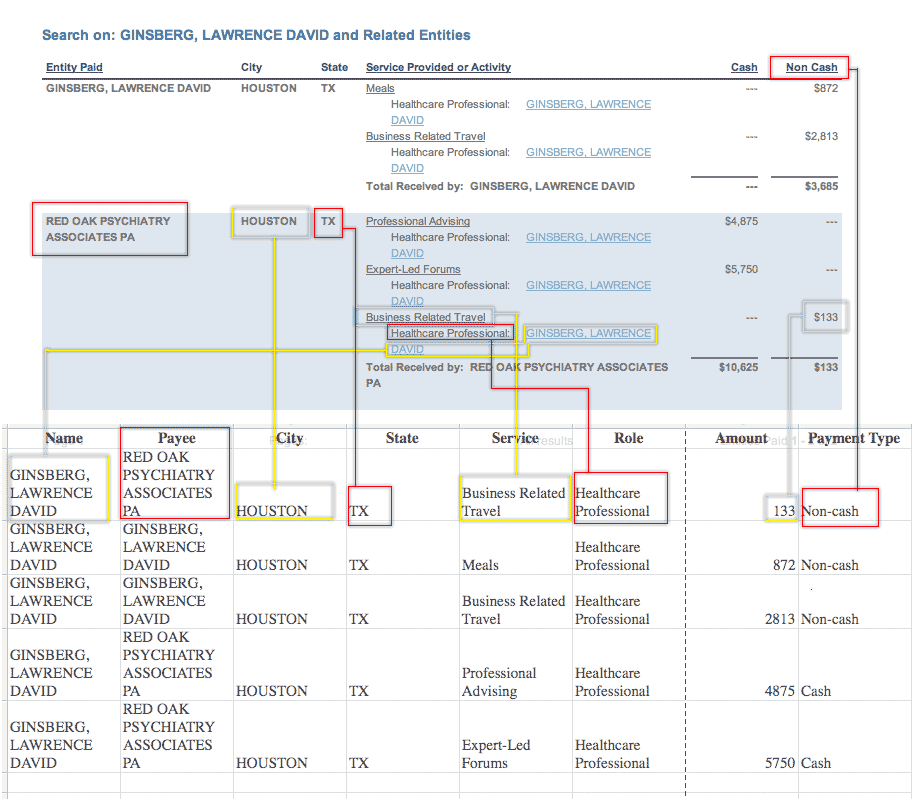
The most important nuance involves how you keep track of payments per category (speaking, consulting, etc.). Other companies’ disclosures (such as Eli Lilly’s) have column headers for each categories, so you know exactly how many types of payment to expect.
But in Pfizer’s case, you don’t know for sure that the above example covers all the possible categories of payment. If you’ve programmed your parser to find a dollar value and place it in either the “Meals”, “Business Related Travel”, “Professional Advising”, or “Forums”, what happens when it reads a page that has a different category, like “Research”?
Rather than design a scraper to add unanticipated columns on the fly, let’s just plan on our compiled textfile to handle categories with two columns, one for the category of service, and one for amount:
Normalized, but not flexible:
| Forums | Travel | Advising | Meals |
|---|---|---|---|
| 872 | |||
| 2813 | |||
| 4875 | |||
| 5750 | |||
| 133 |
Normalized and flexible
| Service | Amount |
|---|---|
| Meals | 872 |
| Business Related Travel | 2813 |
| Professional Advising | 4875 |
| Expert-Led Forums | 5750 |
| Business Related Travel | 133 |
Copying the information from a website is straightforward, but actually parsing HTML that’s meant for easy-readability into normalized data can be an exercise in patience. For example, in Pfizer’s case, each entity-payment-record spans an unpredictable number of rows. The fourth column contains the details of the payments but structures them as lists within lists.
So it takes a little trial-and-error, which is why it’s important to save web pages to your hard drive so you don’t have to re-download them when retrying your parsing code.
Again, we are providing the data we’ve collected for Dollars for Docs, including Pfizer’s disclosures, upon request. The following code is presented as a learning example for Pfizer’s particular data format.
require 'rubygems'
require 'nokogiri'
class String
# a helper function to turn all tabs, carriage returns, nbsp into regular spaces
def astrip
self.gsub(/([\302|\240|\s|\n|\t])|(\ ?){1,}/, ' ').strip
end
end
# We want to keep track of the doc_name to make sure we only grab items with the doctors' name.
# since hcpdisplayName shows ALL doctors who have the same root name, so we want to avoid double counting doc payments
DOC_URL_FILE = "doc-pages-urls.txt"
DOC_PAGES_SUBDIR = 'pfizer-doc-pages'
COMPILED_FILE_NAME = 'all-payments.txt'
compiled_file = File.open(COMPILED_FILE_NAME, 'w')
payment_array = []
Dir.entries(DOC_PAGES_SUBDIR).select{|f| f.match(/\.html/)}.each do |url|
doc_name=url.match(/pfizer-doc-page--(.+?)\.html/)[1]
puts "Reading #{doc_name} page"
page=Nokogiri::HTML(open("#{DOC_PAGES_SUBDIR}/#{url}")).css('#hcpPayments')
# All paid entities are in rows in which the first cell is filled. Each entities associated payments have this first cell
# blank
rows = page.css('tr')
if rows.length > 0
entity_paid,city,state = nil
rows[1..-1].each do |row| # row 0 is the header row, so skip it
# iterate through each row. Rows that have a name in the first cell (entity paid) denote distict entities to which payments went to
cells = row.css('td')
if !cells[0].text.astrip.empty?
# we have a new entity, and city and state here
entity_paid,city,state = cells[0..2].map{|c| c.text.astrip}
end
# the fourth column should always have at least one link that corresponds to the doc_name
doc_link = cells[3].css('a').select{|a| a['href']=="payments_report.jsp?hcpdisplayName=#{doc_name}"}
if doc_link.length == 1
# this is a cell that contains a doctor name and a payment
# it should also contain exactly one link that describe the service provided in a tooltip
service_link = cells[3].css('a').select{|a| a.attributes['onmouseover'] && a.attributes['onmouseover'].value.match(/showTooltip/)}
raise "Not exactly one service link for payee #{entity_paid}: #{url}" if service_link.length != 1
# Now, capture the cash/non-cash cells:
cash,non_cash = cells[4..5]
##
## Write this row to the file
##
compiled_file.puts([entity_paid,city,state, doc_link[0].text, service_link[0].text, cash.text, non_cash.text].map{|t| t.astrip}.join("\t"))
else
## This means that none, or more than one doctor's name was found here
## So the cell was either blank, or it could contain an unexpected name.
## You should write some test conditions here and log the records
## that don't fit your assumptions about the data
end # end of if doc_link.length==1
end # end of rows.each
end
#end of if rows.length > 0
end # end of Dir.entries(DOC_PAGES_SUBDIR).select
compiled_file.close
Scraping Caveats
No matter how well you plan out your data collection, you still might not be able to predict all the idiosyncrasies in the data source. In fact, two basic assumptions I’ve had regarding the Pfizer site weren’t correct:
Each entity paid has an associated doctor – In fact, only some universities only have the amount of research funding, with no names listed. If you’re only interested in doctors with Pfizer connections, this isn’t a huge deal. But you might run into problems if you’ve hard-coded your scraper to expect at least one name there.
The doctor pages only list one doctor – You might assume that a url that reads:
http://www.pfizer.com/responsibility/working_with_hcp/payments_report.jsp?hcpdisplayName=PATEL,+MUKESH
…would belong to a single doctor named “PATEL, MUKESH.” However, it turns out that the hcpdisplayName parameter will retrieve every name that begins with, in this case, “PATEL, MUKESH”. In nearly every case, this returns a page with just one actual doctor. But there are apparently two doctors in Pfizer’s database with that name, “PATEL, MUKESH” and “PATEL, MUKESH MANUBHAI”
So, in the main listing, both PATEL, MUKESH and PATEL, MUKESH MANUBHAI will be listed. Visiting the page with a hcpdisplayName of the shorter-named Patel will bring up the other doctor’s payment records. So to avoid double-counting, you’ll want to program your scraper to only copy the data associated with the exact hcpdisplayName that you’re querying.
It’s possible to set hcpdisplayName to something very broad. ‘F‘, for example, brings up a single page of records for the 166 doctors whose last names start with ‘F’.
So, in your pre-scrape scouting of a website, you should test whether queries require exact parameters or just partial matches. Instead of collecting the 485 pages of 10 doctors each, you could save you and Pfizer some time by collecting the 26 alphabetical lists with the hcpdisplayName.
Once you’re well acquainted with Nokogiri and its powerful HTML parsing methods, you may want to familiarize yourself with mechanize, which uses Nokogiri for parsing, but also provides a convenient set of methods to work with sites that require filling out forms.
The Dollars for Docs Data Guides
Introduction: The Coder’s Cause – Public records gathering as a programming challenge.
- Using Google Refine to Clean Messy Data – Google Refine, which is downloadable software, can quickly sort and reconcile the imperfections in real-world data.
- Reading Data from Flash Sites – Use Firefox’s Firebug plugin to discover and capture raw data sent to your browser.
- Parsing PDFs – Convert made-for-printer documents into usable spreadsheets with third-party sites or command-line utilities and some Ruby scripting.
- Scraping HTML – Write Ruby code to traverse a website and copy the data you need.
- Getting Text Out of an Image-only PDF – Use a specialized graphics library to break apart and analyze each piece of a spreadsheet contained in an image file (such as a scanned document).










